Team Profile
Our aim is research and development concerning the resources related to plants, microorganisms etc., the development of bases for genome information etc. and metabolome analysis approaches, among others. We undertake gene hunt studies related to the enhancement of the production, growth and environmental tolerance of cellulose in Brachypodium, a species of plant expected to serve as the model for studies on the increased production of cellulose biomass. In addition, we perform metagenomics and genomics with the use of single cells in symbiotic microorganisms of termites and other subjects.
Team leader: Kazuo Shinozaki
Research Overview
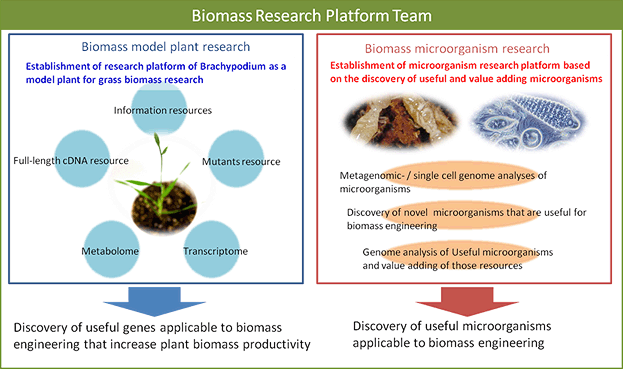
The Biomass Research Platform Team establishes research infrastructure for the promotion of the Biomass Engineering Program, which develops model plants and microorganisms for biomass engineering. For plants, we selected Brachypodium as an emerging model plant for research on grass biomass development. We developed research platforms required for promoting grass biomass research, which include bio-resource (heavy ion beam mutants, full-length cDNA, and ecotypes), advanced omics research (transcriptome and metabolome), and research infrastructure (phenotyping and databases). For microorganisms, we carry out meta-genome and single-cell genome analyses against microorganism communities, such as symbiotic microorganisms observed in termites, to discover key genes for biomass engineering. We also increase value-added discovered microorganisms resources that are beneficial for biomass engineering research.
Research Objective
- Establishment of research platforms for Brachypodium (Brachypodium distachyon), such as large-scale collection of full-length cDNA and generation of mutant resources
- Discovery and application of key genes involved in biomass productivity (yield and stress tolerances) based on reverse genetics and comparative genomics
- Establishment of information resources of plants used as biomass for promoting knowledge integration and exchange
- Meta-genome and single-cell genome analyses against microorganism communities, such as symbiotic microorganisms observed in termites, for discovery of key genes involved in woody biomass degradation, establishment of microorganisms resources beneficial to biomass engineering, and discovery of microorganisms producing bio-plastics
- Analytical advancements of metabolome related to biomass productivity
Brachypodium, an emerging model plant for research on soft cellulose biomass engineering

Brachypodium (Brachypodium distachyon) is an annual grass species found in temperate regions and belongs to the Pooideae subfamily, which includes some cereal species such as wheat and barley. Brachypodium genome draft sequence is available since 2010 and is the first whole genome sequence of a species belonging to the Pooideae subfamily. Brachypodium has many features that make it a model species for research in grass plants: small genome size (272 Mb), small plant size that makes it suitable for cultivation in a small space (laboratory), and high transformation efficiency. Cellulosic biomass of grass species also called as “soft-biomass” can provide high processing suitability because of its lower woody formation than trees. We promote research on discovery of key genes involved in increasing biomass productivity of grass species using Brachypodium.
Establishment of a model plant for biomass engineering research
The Brachypodium genome sequence draft is now available. Immediate establishment of research infrastructures of this emerging model plant is required to accelerate the development of Brachypodium as a model plant for biomass engineering research as well as for molecular breeding of cereal crops. Therefore, we establish Brachypodium bio-resources such as full-length cDNAs and heavy ion beam mutants by using advanced technologies of RIKEN. These activities directly help the worldwide research community involved in Brachypodium research as well as grass biomass and cereal crop research. Furthermore, we also attempt to elucidate cellular systems related to biomass production of grass plants by taking advantage of the advanced technologies for large-scale life science platforms in RIKEN, such as transcriptomics and metabolomics.
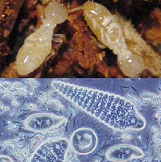
Establishment of a microorganism research platform for efficient biomass utilization
Useful novel genes could be found in genomes of uncultivable microorganisms. We apply meta-genome and single cell genome analysis against uncultivable microorganisms to develop microorganism bio-resources. We promote discovery research as a combinatorial approach with genome analysis to find novel beneficial microorganisms related with polyhydroxyalkanoate biosynthesis, degradation, and fermentation for biomass engineering. Furthermore, we also carry out advanced discovery research to accelerate biomass resource utilization by developing infrastructure, including information resource development.
Research Result
- Construction of full-length cDNA library of Brachypodium
- Sequencing of ca. 40000 clones of Brachypodium full-length cDNAs

